-

-

-
Gregory C Ippolito
B-cell immunology. Antibody repertoires. Vaccines and infectious diseases.-
Dr. Ippolito initially studied physics at Reed College and then completed training in B-cell immunology at The University of Alabama at Birmingham, The University of Cologne (Germany), and The University of Texas at Austin. Key areas of research have focused upon normal and malignant B-cell development (Bcl11a and Foxp1 proto-oncogenes) and the ontogeny of antibody repertoires in health and disease.
RESEARCH SUMMARY
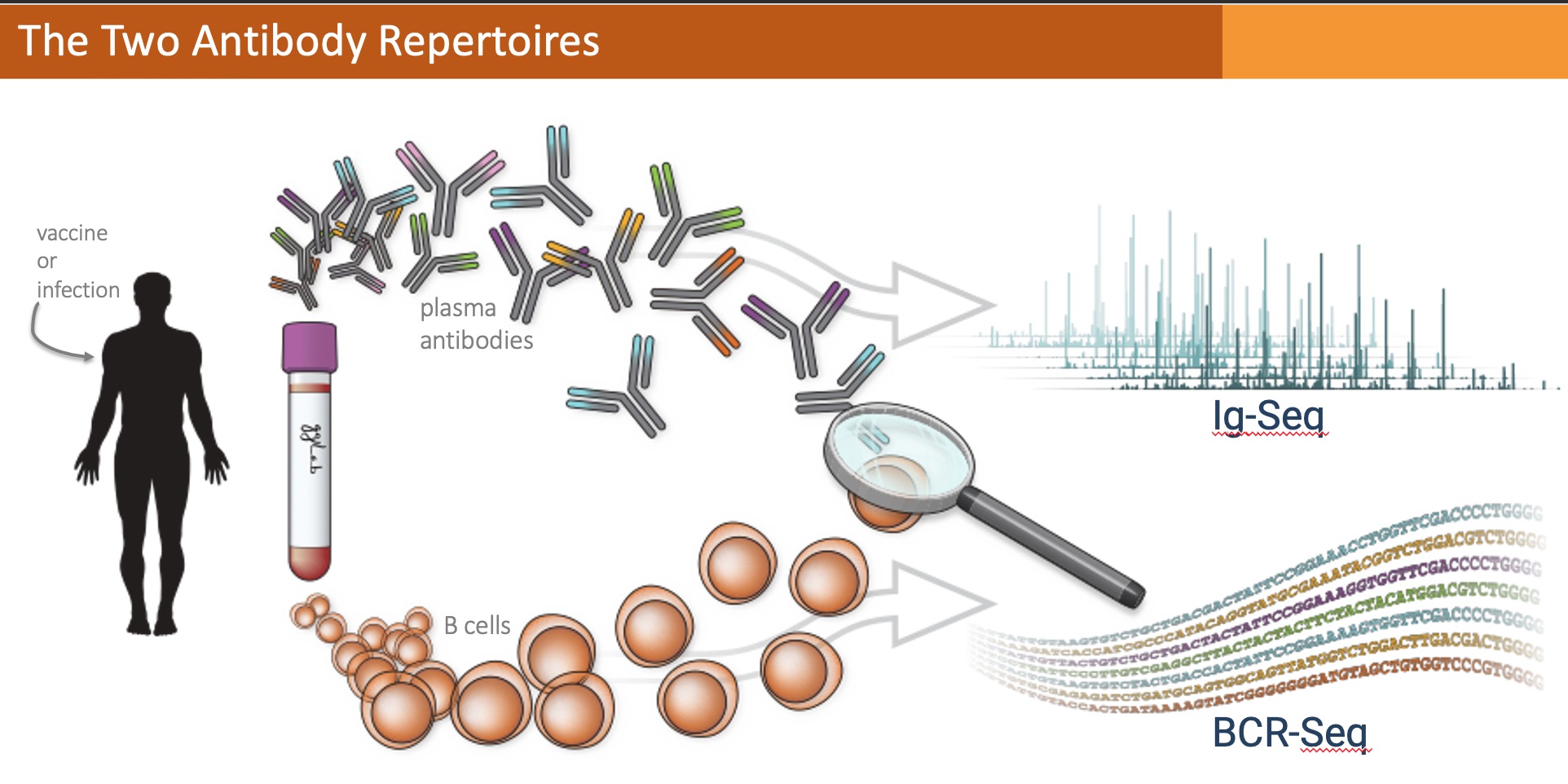
Current research utilizes molecular and proteomic techniques for the comprehensive profiling of antibody repertoires in human adaptive immune responses. The method is a synergistic combination of (i) IgG immunoglobulin protein mass spectrometry and (ii) a high-throughput DNA sequencing method that preserves the natural pairing of B-cell heavy and light chain variable regions (VH:VL regions). This innovation allows for the direct comparison of protein-level plasma IgG immunoglobulin repertoires to DNA-level repertoires in circulating peripheral blood B cells—a unique description that is yielding fresh insights into the temporal dynamics and sequence composition of antibody-mediated immunity. These techniques have been applied to the analysis of vaccine responses, autoimmunity, cancer, and infectious diseases like COVID-19.
Two key technologies, BCR-Seq and Ig-Seq, are used in concert to gather immune response data that is then subjected to bioinformatic analysis.
1. BCR-Seq: Short for “B-cell repertoire sequencing,” this technology allows for analysis of a patient’s immune response at the single-cell level by analyzing the antibody information presented on the surface of the many thousands of individual B-cells produced in response to vaccination or infection.
2. Ig-Seq: Short for “Immunoglobulin sequencing,” this second technology is a proteomic/informatics strategy that, when combined with BCR-Seq data, enables determination of molecular-level identification and functionality of the most prevalent immune response antibody proteins (immunoglobulins) in blood or other fluids.
Current projects
1. Seasonal & Experimental ("Universal") Influenza Vaccines
- NIH Collabortive Influenza Vaccine Innovation Centers (CIVICs) (Duke University)
- NIH Collabortive Influenza Vaccine Innovation Centers (CIVICs) (U. Georgia at Athens/St. Jude's)
- Comparative Analysis of Three Licensed Seasonal Vaccines (CDC/Mayo/Emory)
2. Experimental Blood-Stage Malaria Vaccines
- RH5.1 (PATH/USAID/U. Oxford)
- MSP1, AMA1, RhopH3 (UTHSC San Antonio/Fred Hutch)
3. Infectious Diseases
- COVID-19 NIH SeroNet (U. North Carolina at Chapel Hill)
- COVID-19 Gates Foundation Pan-CoV Vaccines (J. McClellan, UT Austin)
- Dengue Virus NIH (U.C. Berkeley)
4. Autoimmunity
- Systemic lupus erythematosus (SLE) and the DNASE1L3 enzyme (NYU)
- Rheumatoid arthritis (RA) and the PAD4 enzyme (UAB/JHU)
-
2021 Molecular Level Characterization of Circulating Aquaporin-4 Antibodies in Neuromyelitis Optica Spectrum Disorder. Neurol Neuroimmunol Neuroinflamm. 8(5):e1034. *co-corresponding authors
2020 Structure-based design of prefusion-stabilized SARS-CoV-2 spikes, Science 369(6510):1501-1505.
2020 Salazar E, Perez KK, Ashraf M, Chen J, Castillo B, Christensen PA, Eubank T, Bernard DW, Eagar TN, Long SW, Subedi S, Olsen RJ, Leveque C, Schwartz MR, Dey M, Chavez-East C, Rogers J, Shehabeldin A, Joseph D, Williams G, Thomas K, Masud F, Talley C, Dlouhy KG, Lopez BV, Hampton C, Lavinder J, Gollihar JD, Maranhao AC, Ippolito GC, Saavedra MO, Cantu CC, Yerramilli P, Pruitt L, Musser JM. Treatment of COVID-19 Patients with Convalescent Plasma, American Journal of Pathology S0002-9440(20)30257-1.
2020 Soni C, Perez OA, Voss WN, Serpas L, Mehl J, Goike J, Georgiou G, Ippolito GC, Sisirak V, Reizis B. Plasmacytoid dendritic cells promote extrafollicular anti-DNA responses in lupus through type I interferon, Immunity 52(6):1022-1038.
2020 Tanno H, McDaniel JR, Stevens CA, Voss WN, Li J, Durrett R, Lee J, Gollihar J, Tanno Y, Delidakis G, Pothukuchy A, Ellefson JW, Goronzy JJ, Maynard JA, Ellington AD, Ippolito GC, Georgiou G. A facile technology for the high-throughput sequencing of the paired VH:VL and TCRβ:TCRα repertoires, Science Advances eaay9093.
2020 Sterrett S, Peng BJ, Burton RL, LaFon DC, Westfall AO, Singh S, Pride M, Anderson AS, Ippolito GC, Schroeder HW Jr, Nahm MH, Prasad AK, Goepfert P, Bansal A, Peripheral CD4 T follicular cells induced by a conjugated pneumococcal vaccine correlate with enhanced opsonophagocytic antibody responses in younger individuals, Vaccine 38(7):1778-1786.
2019 , , , , , Loss of the FOXP1 Transcription Factor Leads to Deregulation of B Lymphocyte Development and Function at Multiple Stages, ImmunoHorizons
2019 Lindesmith LC, McDaniel JR, Changela A, Verardi R, Kerr SA, Costantini V, Brewer-Jensen PD, Mallory ML, Voss WN, Boutz DR, Blazeck JJ, Ippolito GC, Vinje J, Kwong PD, Georgiou G, Baric RS, Sera Antibody Repertoire Analyses Reveal Mechanisms of Broad and Pandemic Strain Neutralizing Responses after Human Norovirus Vaccination, Immunity 50(6):1530-1541.
2019 Lee J, Paparoditis P, Horton AP, Frühwirth A, McDaniel JR, Jung J, Boutz DR, Hussein DA, Tanno Y, Pappas L, Ippolito GC, Corti D, Lanzavecchia A, Georgiou G, Persistent Antibody Clonotypes Dominate the Serum Response to Influenza over Multiple Years and Repeated Vaccinations, Cell Host & Microbe 25(3):367-376.e5.
2018 Manso TC, Groenner-Penna M, Minozzo JC, Antunes BC, Ippolito GC, Molina F, Felicori LF, Next-generation sequencing reveals new insights about gene usage and CDR-H3 composition in the horse antibody repertoire, Mol Immunol 105:251-259.
2018 Johnson EL, Doria-Rose NA, Gorman J, Bhiman JN, Schramm CA, Vu AQ, Law WH, Zhang B, Bekker V, Abdool Karim SS, Ippolito GC, Morris L, Moore PL, Kwong PD, Mascola JR, Georgiou G, Sequencing HIV-neutralizing antibody exons and introns reveals detailed aspects of lineage maturation, Nature Communications 9(1):4136.
2018 Peiris H, Park S, Louis S, Gu X, Lam JY, Asplund O, Ippolito GC, Bottino R, Groop L, Tucker H, Kim SK, Discovering human diabetes-risk gene function with genetics and physiological assays, Nature Communications 9(1):3855.
2018 , , , , , , , , ,
2018 Liu Y, McDaniel JR, Khan S, Campisi P, Propst EJ, Holler T, Grunebaum E, Georgiou G, Ippolito GC, Ehrhardt GRA, Antibodies Encoded by FCRL4-Bearing Memory B Cells Preferentially Recognize Commensal Microbial Antigens, J Immunol doi: 10.4049/jimmunol.1701549.
2018 Sharma R, Al‐Saleem FH, Panzer J , Lee J, Puligedda RD, Felicori LF, Kattala, CD, Rattelle AJ, Ippolito GC, Cox RH, Lynch DR, and Dessain SK, Monoclonal antibodies from a patient with anti‐NMDA receptor encephalitis, Ann Clin Transl Neurol doi:10.1002/acn3.592
2018 McDaniel JR, Pero SC, Voss WN, Shukla GS, Sun Y, Schaetzle S, Lee CH, Horton AP, Harlow S, Gollihar J, Ellefson JW, Krag CC, Tanno Y, Sidiropoulos N, Georgiou G, Ippolito GC*, Krag DN, Identification of tumor-reactive B cells and systemic IgG in breast cancer based on clonal frequency in the sentinel lymph node, Cancer Immunol Immunother doi: 10.1007/s00262-018-2123-2. *co-corresponding author
2017 McDaniel JR, Ippolito GC, Georgiou G, Mapping the secrets of the antibody pool, Nature Biotechnology doi:10.1038/nbt.3972
2017 Tanaka T, Zhang W, Sun Y, Shuai Z, Chida A, Kenny TP, Yang G-X, Sanz I, Ansai A, Bowlus CL, Ippolito GC, Coppel RL, Okazaki K, He X-S, Leung PSC, Gershwin ME, Autoreactive Monoclonal Antibodies from Patients with Primary Biliary Cholangitis Recognize Environmental Xenobiotics, Hepatology doi: 10.1002/hep.29245.
2017 Lau D, Lan LY, Andrews SF, Henry C, Rojas KT, Neu KE, Huang M, Huang Y, DeKosky B, Palm A-KE, Ippolito GC, Georgiou G, Wilson PC, Low CD21 expression defines a population of recent germinal center graduates primed for plasma cell differentiation, Science Immunology doi: 10.1126/sciimmunol.aai8153.
2016 Wu GC, Cheung N-KV, Georgiou G, Marcotte EM, Ippolito GC, Temporal stability and molecular persistence of the bone marrow plasma cell antibody repertoire, Nature Communications doi: 10.1038/ncomms13838.
2016 Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee C-H, Lavinder JJ, Murrin EM, Chrysostomou C, Hoi KH, Tsybovsky Y, Thomas PV, Druz A, Zhang B, Zhang Y, Wang L, Kong W-P, Park D, Popova LI, Dekker CL, Davis MM, Bloom CE, Ross TM, Ellington AD, Wilson PC, Marcotte EM, Mascola JR, Ippolito GC, Krammer F, Quake SR, Kwong PD, Georgiou G, Quantitative, molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination, Nature Medicine doi:10.1038/nm.4224.
2016 Raymond DD, Stewart SM, Lee J, Ferdman J, Bajic G, Do KT, Ernandes MJ, Suphaphiphat P, Settembre EC, Dormitzer PR, Del Giudice G, Finco O, Kang TH, Ippolito GC, Georgiou G, Kepler TB, Haynes BF, Moody MA, Liao HX, Schmidt AG, Harrison SC, Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nature Medicine 22(12):1465-1469.
2016 Steiniger SCJ, Glanville J, Harris DW, Wilson TL, Ippolito GC, Dunham SA, Comparative analysis of the feline immunoglobulin repertoire, Biologicals 46: 81–87.
2016 Woodworth MB, Greig LC, Liu KX, Ippolito GC, Tucker HO, Macklis JD, Ctip1 Regulates the Balance between Specification of Distinct Projection Neuron Subtypes in Deep Cortical Layers Regulates the Balance between Specification of Distinct Projection Neuron Subtypes in Deep Cortical Layers, Cell Rep S2211-1247(16)30335-7. doi: 10.1016/j.celrep.2016.03.064
2016 DeKosky BJ, Lungu OI, Park D, Johnson EL, Charab W, Chrysostomou C, Kuroda D, Ellington AD, Ippolito GC, Gray JJ, Georgiou G, Large-Scale Sequence and Structural Comparisons of Human Naïve and Antigen-Experienced Antibody Repertoires, Proc Natl Acad Sci USA doi: 10.1073/pnas.1525510113
2016 Dekker JD, Park D, Shaffer AL III, Kohlhammer H, Deng W, Lee B-K, Ippolito GC, Georgiou G, Iyer VR, Staudt LM, Tucker HO, Subtype Specific Addiction of the Activated B Cell Subset of Diffuse Large B Cell Lymphoma to FOXP1, Proc Natl Acad Sci USA doi: 10.1073/pnas.1524677113
2015 Wine Y, Horton AP, Ippolito GC, Georgiou G, Serology in the 21st century: the molecular-level analysis of the serum antibody repertoire, Curr Opin Immunol 35:89-97.
2015 Silva-Sanchez A, Liu CR, Vale AM, Khass M, Kapoor P, Elgavish A, Ivanov II, Ippolito GC, Schelonka RL, Schoeb TR, Burrows PD, Schroeder HW Jr., Violation of an Evolutionarily Conserved Immunoglobulin Diversity Gene Sequence Preference Promotes Production of dsDNA-Specific IgG Antibodies, PLoS ONE 10(2):e0118171
2015 Lavinder JJ, Horton AP, Georgiou G, Ippolito GC, Next-generation sequencing and protein mass spectrometry for the comprehensive analysis of human cellular and serum antibody repertoires, Current Opinion in Chemical Biology 24C:112-120.
2015 DeKosky BJ, Kojima T, Rodin A, Charab W, Ippolito GC, Ellington AD, Georgiou G, In-Depth Determination and Analysis of the Human Paired Heavy and Light Chain Antibody Repertoire, Nature Medicine 21(1):86-91
2014 Szymanska Mroczek E, Ippolito GC, Rogosch T, Hoi K-H, Hwangpo TA, Brand MG, Zhuang Y, Liu CR, Schneider DA, Zemlin M, Brown E, Georgiou G, Schroeder HW Jr, Differences in the composition of the human antibody repertoire by B cell subsets in the blood, Front Immunol 5:96
2014 Ippolito GC*, Dekker JD*, Wang Y-H, Lee B-K, Shaffer AL III, Lin J, Wall JK, Lee B-S, Staudt LM, Liu Y-J, Iyer VR, Tucker HO, Dendritic cell fate is determined by BCL11A, Proc Natl Acad Sci USA (*co-first authors) doi:10.1073/pnas.1319228111
2014 Steiniger SCJ, Dunkle WE, Bammert GF, Wilson TL, Krishnan A, Dunham SA, Ippolito GC, Bainbridge G, Fundamental characteristics of the expressed immunoglobulin VH and VL repertoire in different canine breeds in comparison with those of humans and mice, Mol Immunology 59: 71-78
2014 Lavinder J, Wine Y, Giesecke C, Ippolito GC, Horton AP, Hoi K-H, DeKosky BJ, Ellington AD, Dörner T, Marcotte EM, Boutz DR, Georgiou G, Identification and characterization of the constituent human serum antibodies elicited by vaccination, Proc Natl Acad Sci USA 111(6):2259-64 doi/10.1073/pnas.1317793111
2014 Georgiou G, Ippolito GC, Beausang J, Busse CE, Wardemann H, Quake SR, The promise and challenge of high-throughput sequencing of the antibody repertoire, Nat Biotechnol 32(2): 1-11
2013 Hoi KH, Ippolito GC, Intrinsic bias and public rearrangements in the human immunoglobulin Vλ light chain repertoire, Genes Immunity 14(4):271-6
2013 Lee BS, Dekker JD, Lee BK, Iyer VR, Sleckman BP, Shaffer AL 3rd, Ippolito GC, Tucker PW, The BCL11A transcription factor directly activates RAG gene expression and V(D)J recombination, Mol Cell Biol 33(9):1768-81
2013 DeKosky BJ, Ippolito GC, Deschner RP, Lavinder JJ, Wine Y, Rawlings BM, Varadarajan N, Giesecke C, Dörner T, Andrews SF, Wilson PC, Hunicke-Smith SP, Willson CG, Ellington AD, Georgiou G, High-throughput sequencing of the paired human immunoglobulin heavy and light chain repertoire, Nat Biotechnol 31(2):166-92012 Rogosch T, Kerzel S, Hoi KH, Zhang Z, Maier RF, Ippolito GC, Zemlin M, Immunoglobulin analysis tool: a novel tool for the analysis of human and mouse heavy and light chain transcripts, Front Immunol 3:176
2012 Ippolito GC, Hoi KH, Reddy ST, Carroll SM, Ge X, Rogosch T, Zemlin M, Shultz LD, Ellington AD, Vandenberg CL, Georgiou G, Antibody repertoires in humanized NOD-scid-IL2Rγ(null) mice and human B cells reveals human-like diversification and tolerance checkpoints in the mouse, PLoS ONE 7(4):e35497
2011 Sürmeli G, Akay T, Ippolito GC, Tucker PW, Jessell ™, Patterns of spinal sensory-motor connectivity prescribed by a dorsoventral positional template, Cell 147(3):653-65
2011 Xu J, Peng C, Sankaran VG, Shao Z, Esrick EB, Chong BG, Ippolito GC, Fujiwara Y, Ebert BL, Tucker PW, Orkin SH, Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing, Science 334(6058):993-62010 Feng X, Ippolito GC, Tian L, Wiehagen K, Oh S, Sambandam A, Willen J, Bunte RM, Maika SD, Harriss JV, Caton AJ, Bhandoola A, Tucker PW, Hu H, Foxp1 is an essential transcriptional regulator for the generation of quiescent naive T cells during thymocyte development, Blood 115(3):510-8
2009 Sankaran VG, Xu J, Ragoczy T, Ippolito GC, Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, Tucker PW, Orkin SH, Developmental and species-divergent globin switching are driven by BCL11A, Nature 460(7259):1093-72009 Schmidt C, Kim D, Ippolito GC, Naqvi HR, Probst L, Mathur S, Rosas-Acosta G, Wilson VG, Oldham AL, Poenie M, Webb CF, Tucker PW, Signalling of the BCR is regulated by a lipid rafts-localised transcription factor, Bright, EMBO J 28(6):711-24
2008 Zemlin M, Schelonka RL, Ippolito GC, Zemlin C, Zhuang Y, Gartland GL, Nitschke L, Pelkonen J, Rajewsky K, Schroeder HW Jr, Regulation of repertoire development through genetic control of DH reading frame preference, J Immunol 181(12):8416-24
2007 Koon HB, Ippolito GC, Banham AH, Tucker PW, FOXP1: a potential therapeutic target in cancer, Expert Opin Ther Targets 11(7):955-65
2007 Lin D, Ippolito GC, Zong RT, Bryant J, Koslovsky J, Tucker P. Bright/ARID3A contributes to chromatin accessibility of the immunoglobulin heavy chain enhancer, Mol Cancer 26;6:23
2006 Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, Rajewsky K, Schroeder HW Jr, Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production, J Exp Med 203(6):1567-78
2006 Pulford K, Banham AH, Lyne L, Jones M, Ippolito GC, Liu H, Tucker PW, Roncador G, Lucas E, Ashe S, Stockwin L, Walewska R, Karran L, Gascoyne RD, Mason DY, Dyer MJ, The BCL11AXL transcription factor: its distribution in normal and malignant tissues and use as a marker for plasmacytoid dendritic cells, Leukemia. 2006 Aug;20(8):1439-41.
2006 Liu H, Ippolito GC, Wall JK, Niu T, Probst L, Lee BS, Pulford K, Banham AH, Stockwin L, Shaffer AL, Staudt LM, Das C, Dyer MJ, Tucker PW, Functional studies of BCL11A: characterization of the conserved BCL11A-XL splice variant and its interaction with BCL6 in nuclear paraspeckles of germinal center B cells, Mol Cancer 5:18
-
BIO 360K/394L ADVANCED IMMUNOLOGY
(Spring semesters 2020–2022)
-
Graduate/Post Doc Opportunities
Our lab is always looking for talented and dedicated grad students and postdocs. If you find the research that we do interesting and would like to hear more, please send an email to Greg at gci[at]utexas.edu.
-