Antibiotics
-
THIS STREAM IS NO Longer Active.
Full Stream Name: Antibiotics: Discovery and Function
Principal Investigator: Bryan Davies
Can we discover new antibiotics, and what effects do antibiotic-producing bacteria have on other members of a microbial community?
As the numbers of antibiotic-resistant bacterial strains like methicillin-resistant Staphylococcus aureus (MRSA) are increasing, we are rapidly running out of last line drugs to treat infections. Nearly all of the antibiotics currently used in hospitals and clinics come from microorganisms that are commonly found in the soil. Students in this stream will isolate new antibiotic-producing bacteria from the soil right here on campus, determine the chemical nature of the drug(s) they produce, and use next generation sequencing technology to find out how the presence of antibiotic producers affects other bacteria in the community. At the stream’s conclusion it is possible that students will have both discovered a new antibiotic and determined its role in shaping the soil microbiome.Background:
The numbers of antibiotic-resistant strains of bacteria are rising, and over 20,000 genes that promote antibiotic resistance across a range of pathogens have been described. It is critical that researchers identify novel antibiotics in order to combat these resistant bugs. Approximately 60% of drugs developed in the last 20 years are derived from natural products, and nearly every antibiotic that we currently use was first discovered in a microorganism. Among these microorganisms is a family of bacteria called Streptomyces that produces several antibiotics including streptomycin, which is the basis of several antibiotics used in hospitals and clinics today. While the ability of these bacteria to produce antibiotics is useful to humans seeking new therapeutics, the role these organisms play in the soil community and the reasons for their production of antibiotics remain unclear. Watch the stream video.
Research Questions:
1. Can we discover new kinds of antibiotics?
Nearly all of the antibiotics currently used in clinics and hospitals were originally discovered as natural products made by bacteria and fungi that live in the soil. The first objective of the Antibiotics stream is to isolate and characterize new, antibiotic-producing soil bacteria and determine the nature of the antibiotics they produce.
2. How do antibiotic producers affect other soil bacteria?
While microbiologists have known of the existence of antibiotic-producing soil bacteria for many years, the role these organisms play within the soil microbial community is not well understood. It seems logical that antibiotic production evolved as a means to compete with other soil bacteria, but many antibiotic producers do not generate high enough levels of the drugs to kill even their closest neighbors.
Experimental Techniques:
Microbiology
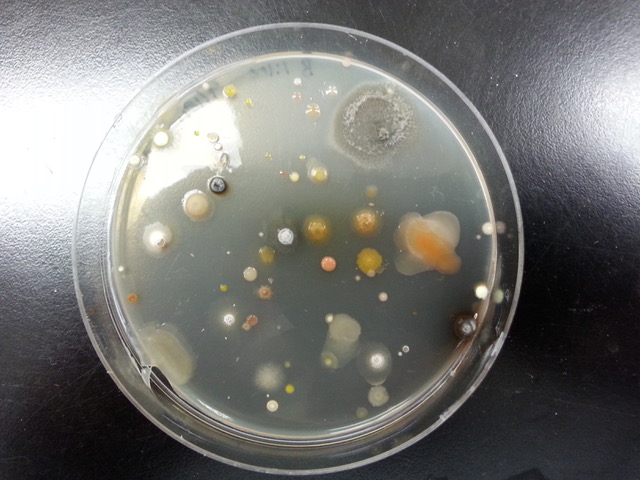
The first step for students in the Antibiotics stream is to isolate antibiotic-producing soil bacteria. This process involves collecting soil samples, spreading the soil on agar plates containing a growth medium that enriches for antibiotic producers, and purifying a single bacterial strain from all the bacteria on the plate. Once an individual strain has been isolated, the next step is to use physical and biochemical methods to characterize the organism. By determining the biochemical properties of a bacterium, students can both identify the organism and begin to determine its role within the soil microbial community.
Chemistry
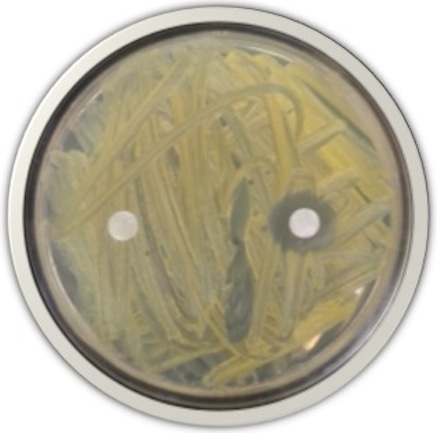
Once an antibiotic producer has been identified, students extract its antibiotics using organic solvents. The extracts will then be subjected to separation techniques like Thin Layer Chromatography (TLC) and High Performance Liquid Chromatography (HPLC) to purify the antibiotic of interest. The molecular nature of purified antibiotics is determined, and pharmacological testing is conducted to determine the antibiotic’s efficacy against other bacteria. Students may also use the antibiotic to cure plant infections.
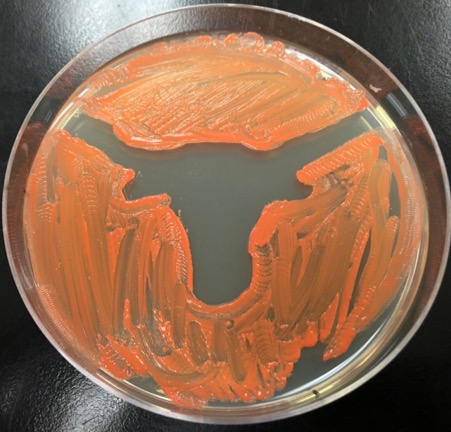
Molecular Biology
Students use several different molecular biology techniques throughout the stream. Molecular characterization of isolated bacteria involves amplification of a small part of the bacterium’s genome using polymerase chain reaction (PCR). The genome fragment will then be sequenced and compared to online databases of previously identified bacteria. If no matching sequence is found, it is likely a new strain has been discovered! Students may also sequence their bacterium’s entire genome using the same technology that is rapidly being adapted for use in hospitals to diagnose disease and identify new treatments. Knowing what genes are present in the genome helps students determine their isolate’s genetic toolkit and potentially which kinds of antibiotics it produces.
-
 No
No - Biochemistry, Biology



